GeMoMa
by Jens Keilwagen, Michael Wenk, Jessica L. Erickson, Martin H. Schattat, Jan Grau, and Frank Hartung
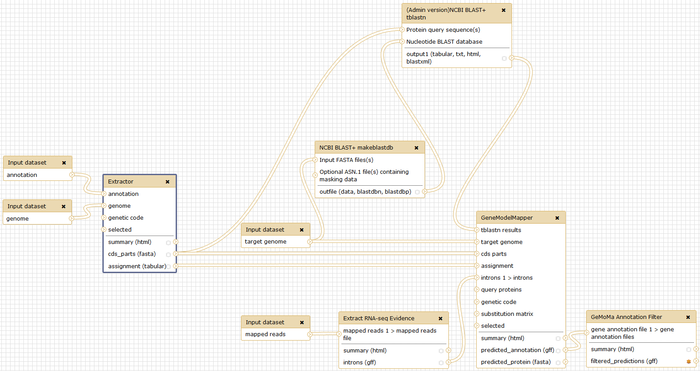
Gene Model Mapper (GeMoMa) is a homology-based gene prediction program that uses the annotation of protein-coding genes in a reference genome utilizing amino acid and intron position conservation to infer annotation of protein-coding genes in a target genome.
Abstract
Annotation of protein-coding genes is very important in many fields of research and application. Thereby, homology-based gene prediction programs allow for transferring knowledge from an annotated organism to an organism of interest.
Here, we present a homology-based gene prediction program called GeMoMa. GeMoMa utilizes the intron position conservation of related genes in different organisms. We assess the performance of GeMoMa comparing it with state-of-the-art competitors on plant and animal genomes using an extended best reciprocal hit approach. We find that it often makes more precise predictions than its competitors yielding an improvement of up to 622% more correct transcripts. Subsequently, we use RNA-seq data to compare the predictions of homology-based gene prediction programs, and find again that GeMoMa performs well.
Hence, we conclude that exploiting intron position conservation improves homology-based gene prediction, and we make GeMoMa freely available as command-line tool and Galaxy integration.
Paper
If you use GeMoMa, please cite
J. Keilwagen, M. Wenk, J. L. Erickson, M. H. Schattat, J. Grau, and F. Hartung. Using intron position conservation for homology-based gene prediction. Nucleic Acids Research, 2016. doi: 10.1093/nar/gkw092
Galaxy
GeMoMa is available in a public web-server at galaxy.informatik.uni-halle.de. The provided web-server only allows a limited number of reference genes and uses a time out of 2 minutes per transcript prediction. For unlimited use, please use the command line program or integrate GeMoMa in your only Galaxy instance.
Download
GeMoMa is implemented in Java using Jstacs. You can download a zip file containing a readme, the GeMoMa jar file and some tiny scripts for running GeMoMa. The jar file allows for
- running the command line interface (CLI) version
- creating the XML file needed for the Galaxy integration.
Running the command line application
For running the command line application, Java v1.8 or later is required.
Extract RNA-seq Evidence (ERE)
For post-processing the mapped RNA-seq data, we provide the tool ExtractRNAseqEvidence (ERE). You can run Extractor from the command line with
java -jar GeMoMa-1.4.jar CLI ERE [<parameter>=<value> ...]
The parameters comprise:
| name | comment | type |
| s | Stranded (Defines whether the reads are stranded. In case of FR_FIRST_STRAND, the first read of a read pair or the only read in case of single-end data is assumed to be located on forward strand of the cDNA, i.e., reverse to the mRNA orientation. If you are using Illumina TruSeq you should use FR_FIRST_STRAND., range={FR_UNSTRANDED, FR_FIRST_STRAND, FR_SECOND_STRAND}, default = FR_UNSTRANDED) | STRING |
| m | mapped reads file (BAM/SAM files containing the mapped reads) | FILE |
| v | ValidationStringency (Defines how strict to be when reading a SAM or BAM, beyond bare minimum validation., range={STRICT, LENIENT, SILENT}, default = LENIENT) | STRING |
| u | use secondary alignments (allows to filter flags in the SAM or BAM, default = true) | BOOLEAN |
| c | coverage output (allows to output the coverage, default = false) | BOOLEAN |
| outdir | The output directory, defaults to the current working directory (.) | STRING |
Extractor
For preparing the data, we provide the tool Extractor. You can run Extractor from the command line with
java -jar GeMoMa-1.4.jar CLI Extractor [<parameter>=<value> ...]
The parameters comprise:
| name | comment | type |
| a | annotation (Reference annotation file (GFF or GTF), which contains gene models annotated in the reference genome) | FILE |
| g | genome (Reference genome file (FASTA)) | FILE |
| gc | genetic code (optional user-specified genetic code, OPTIONAL) | FILE |
| p | proteins (whether the complete proteins sequences should returned as output, default = false) | BOOLEAN |
| c | cds (whether the complete CDSs should returned as output, default = false) | BOOLEAN |
| r | repair (if a transcript annotation can not be parsed, the program will try to infer the phase of the CDS parts to repair the annotation, default = false) | BOOLEAN |
| s | selected (The path to list file, which allows to make only a predictions for the contained transcript ids. The first column should contain transcript IDs as given in the annotation. Remaining columns will be ignored., OPTIONAL) | FILE |
| Ambiguity | Ambiguity (This parameter defines how to deal with ambiguities in the DNA. There are 3 options: EXCEPTION, which will remove the corresponding transcript, AMBIGUOUS, which will use an X for the corresponding amino acid, and RANDOM, which will randomly select an amnio acid from the list of possibilities., range={EXCEPTION, AMBIGUOUS, RANDOM}, default = EXCEPTION) | STRING |
| f | full-length (A flag which allows for choosing between only full-length and all (i.e., full-length and partial) transcripts, default = true) | BOOLEAN |
| v | verbose (A flag which allows to output wealth of additional information, default = false) | BOOLEAN |
| outdir | The output directory, defaults to the current working directory (.) | STRING |
Gene Model Mapper (GeMoMa)
For predicting gene models, we provide the tool GeMoMa. You can run GeMoMa from the command line with
java -jar GeMoMa-1.4.jar CLI GeMoMa [<parameter>=<value> ...]
The parameters comprise:
| name | comment | type | ||||||||||||||||||
| t | tblastn results (The sorted tblastn results) | FILE | ||||||||||||||||||
| tg | target genome (The target genome file (FASTA), i.e., the target sequences in the blast run. Should be in IUPAC code) | FILE | ||||||||||||||||||
| c | cds parts (The query cds parts file (FASTA), i.e., the cds parts that have been blasted) | FILE | ||||||||||||||||||
| a | assignment (The assignment file, which combines parts of the CDS to transcripts, OPTIONAL) | FILE | ||||||||||||||||||
| i | introns (Introns (GFF), which might be obtained from RNAseq, OPTIONAL) | FILE | ||||||||||||||||||
| r | reads (if introns are given by a GFF, only use those which have at least this number of supporting split reads, valid range = [1, 2147483647], default = 1) | INT | ||||||||||||||||||
| s | splice (if no intron is given by RNAseq, compute candidate splice sites or not, default = true) | BOOLEAN | ||||||||||||||||||
| coverage | coverage (experimental coverage (RNAseq), range={NO, UNSTRANDED, STRANDED}, default = NO)
| |||||||||||||||||||
| q | query proteins (optional query protein file (FASTA) for computing the optimal alignment score against complete protein prediction, OPTIONAL) | FILE | ||||||||||||||||||
| g | genetic code (optional user-specified genetic code, OPTIONAL) | FILE | ||||||||||||||||||
| sm | substitution matrix (optional user-specified substitution matrix, OPTIONAL) | FILE | ||||||||||||||||||
| go | gap opening (The gap opening cost in the alignment, default = 11) | INT | ||||||||||||||||||
| ge | gap extension (The gap extension cost in the alignment, default = 1) | INT | ||||||||||||||||||
| m | maximum intron length (The maximum length of an intron, default = 15000) | INT | ||||||||||||||||||
| intron-loss-gain-penalty | intron-loss-gain-penalty (The penalty used for intron loss and gain, default = 25) | INT | ||||||||||||||||||
| e | e-value (The e-value for filtering blast results, default = 100.0) | DOUBLE | ||||||||||||||||||
| ct | contig threshold (The threshold for evaluating contigs, valid range = [0.0, 1.0], default = 0.4) | DOUBLE | ||||||||||||||||||
| rt | region threshold (The threshold for evaluating regions, valid range = [0.0, 1.0], default = 0.9) | DOUBLE | ||||||||||||||||||
| h | hit threshold (The threshold for adding additional hits, valid range = [0.0, 1.0], default = 0.9) | DOUBLE | ||||||||||||||||||
| p | predictions (The (maximal) number of predictions per transcript, default = 10) | INT | ||||||||||||||||||
| selected | selected (The path to list file, which allows to make only a predictions for the contained transcript ids. The first column should contain transcript IDs as given in the annotation. Remaining columns can be used to determine a target region that should be overlapped by the prediction, if columns 2 to 5 contain chromosome, strand, start and end of region, OPTIONAL) | FILE | ||||||||||||||||||
| as | avoid stop (A flag which allows to avoid stop codons in a transcript (except the last AS), default = true) | BOOLEAN | ||||||||||||||||||
| approx | approx (whether an approximation is used to compute the score for intron gain, default = true) | BOOLEAN | ||||||||||||||||||
| align | align (A flag which allows to output a tab-delimited file, which contains the results in a blast-like format (deprecated), default = false) | BOOLEAN | ||||||||||||||||||
| genomic | genomic (A flag which allows to output a fasta file containing the genomic regions of the predictions, default = false) | BOOLEAN | ||||||||||||||||||
| prefix | prefix (A prefix to be used for naming the predictions, default = ) | STRING | ||||||||||||||||||
| tag | tag (A user-specified tag for transcript predictions in the third column of the returned gff. It might be beneficial to set this to a specific value for some genome browsers., default = prediction) | STRING | ||||||||||||||||||
| v | verbose (A flag which allows to output wealth of additional information per transcript, default = false) | BOOLEAN | ||||||||||||||||||
| timeout | timeout (The (maximal) number of seconds to be used for the predictions of one transcript, if exceeded GeMoMa does not ouput a prediction for this transcript., valid range = [0, 604800], default = 3600) | LONG | ||||||||||||||||||
| outdir | The output directory, defaults to the current working directory (.) | STRING | ||||||||||||||||||
GeMoMa returns the predicted annotation as gff file and the predicted proteins as fasta file.
GeMoMa Annotation Filter (GAF)
For predicting gene models, we provide the tool GAF. You can run GAF from the command line with
java -jar GeMoMa-1.4.jar CLI GAF [<parameter>=<value> ...]
The parameters comprise:
| name | comment | type |
| t | tag (the tag used to read the GeMoMa annotations, default = prediction) | STRING |
| r | relative score filter (the initial filter on the relative score (i.e. score devided by length), default = 0.75) | DOUBLE |
| c | complete (only complete predictions (having start and stop codon) pass the initial filter, default = true) | BOOLEAN |
| m | missing intron evidence filter (the filter for single-exon transcripts or if no RNA-seq data is used, decides for overlapping other transcripts whether they should be used (=true) or discarded (=false), default = false) | BOOLEAN |
| i | intron evidence filter (the filter on the intron evidence given by RNA-seq-data for overlapping transcripts, valid range = [0.0, 1.0], default = 1.0) | DOUBLE |
| cbf | common border filter (the filter on the common borders of transcripts, the lower the more transcripts will be checked as alternative splice isoforms, valid range = [0.0, 1.0], default = 0.75) | DOUBLE |
| g | gene annotation files (GFF files containing the gene annotations (predicted by GeMoMa)) | FILE |
| e | evidence percentage filter (Each gene annotation file is handle as separate evidence. A prediction is only returned if it is contained at least in this percentage of evidence files.), valid range = [0.0, 1.0], default = 0.5) | DOUBLE |
| outdir | The output directory, defaults to the current working directory (.) | STRING |
CompareTranscripts
For comparing gene models from GeMoMa predictions with existing annotation, we provide the tool CompareTranscripts. You can run CompareTranscripts from the command line with
java -jar GeMoMa-1.4.jar CLI CompareTranscripts [<parameter>=<value> ...]
The parameters comprise:
| name | comment | type |
| p | prediction (The predicted annotation) | FILE |
| a | annotation (The true annotation) | FILE |
| assignment | assignment (the transcript info for the reference of the prediction, OPTIONAL) | FILE |
| outdir | The output directory, defaults to the current working directory (.) | STRING |
AnnotationEvidence
For providing RNA-seq evidence (e.g. tie) for existing annotation, we provide the tool AnnotationEvidence. You can run AnnotationEvidence from the command line with
java -jar GeMoMa-1.4.jar CLI AnnotationEvidence [<parameter>=<value> ...]
The parameters comprise:
| name | comment | type | ||||||||||||||||||
| a | annotation (The genome annotaion file (GFF)) | FILE | ||||||||||||||||||
| g | genome (The genome file (FASTA), i.e., the target sequences in the blast run. Should be in IUPAC code) | FILE | ||||||||||||||||||
| i | introns file (Introns (GFF), which might be obtained from RNAseq, OPTIONAL) | FILE | ||||||||||||||||||
| r | reads (if introns are given by a GFF, only use those which have at least this number of supporting split reads, valid range = [1, 2147483647], default = 1) | INT | ||||||||||||||||||
| c | coverage file (experimental coverage (RNAseq), range={NO, UNSTRANDED, STRANDED}, default = NO)
| |||||||||||||||||||
| outdir | The output directory, defaults to the current working directory (.) | STRING | ||||||||||||||||||
FAQs
- Why does the Extractor not return a single CDS-part, protein, ...?
- Please check whether the names of your contigs/chromosomes in your annotation (gff) and genome file (fasta) are identical. The fasta comments should at best only contain the contig/chromosome name. (Since GeMoMa 1.4, comments, which contain the contig/chromosome name and some additional information separated by a space, are also fine.) In addition, check the statistics that are given by the Extractor. It lists how many genes have been read and how many genes have been removed for different reasons. One common problem is that some annotation files do not include the stop codon in the CDS annotation.
- How can I force GeMoMa to make more predictions?
- There are several parameters affecting the number of predictions. The most prominent are the number of predictions (p) and the contig threshold (ct). For each reference transcript/CDS, GeMoMa initially makes a preliminary prediction and uses this prediction to determine whether a contig is promising and should be used to determine the final predictions. You may decrease ct and increase p to have more contigs in the final prediction. Increasing the number of predictions allows GeMoMa to output more predictions that have been computed. Decreasing the contig threshold allows to increase the number of predictions that are (internally) computed. Increasing p to a very large number without decreasing ct does not help.
- Running GeMoMa on a single contig of my assembly yield thousands of weird predictions. What went wrong?
- By default, GeMoMa is not build to be run on a single contig. GeMoMa tries to make predictions for all given reference CDS in the given target sequence(s). If the given target sequence is only a fraction of the complete target genome/assembly, GeMoMa will produce weird predictions as it does not filter for the quality of the predictions internally. There are two options to handle this:
- Use a list of gene models that you expect to be located on this contig (cf. parameter "selected").
- Filter the predictions using GAF (cf.
java -jar GeMoMa-<version>.jar CLI GAF).
- Is it mandatory to use RNA-seq data?
- No, GeMoMa is able to make predictions with and without RNA-seq evidence.
- Is it possible to use multiple reference organisms?
- It is possible to use multiple reference organisms for GeMoMa. Just run GeMoMa on each reference organism separately. Finally, you can employ GAF (cf.
java -jar GeMoMa-<version>.jar CLI GAF) to combine these annotations. - Why do some reference genes not lead to a prediction in the target genome?
- Please first check whether your reference genes have been discarded by the Extractor (cf. assignment file).
- If the genes have been discarded, there are two possibilities:
- The CDS might be redundant, i.e. the coding exons are identical to those of another transcript. In this case, only one CDS is further evaluated.
- There might be something wrong with your reference genes, e.g., missing start codon, missing stop codon, premature stop codon, ambiguous nucleotides, ... and you should check the options of Extractor or the annotation.
- If the reference genes passed the Extractor, there are several possible explanations for this behavior. The two most prominent are:
- GeMoMa stopped the prediction of a reference genes since it does not return a result within the given time (cf. parameter "timeout").
- GeMoMa simply did not find a prediction matching the remaining quality criteria.
- For two different reference transcripts, the predictions of GeMoMa overlap or are identical. What should I do with those?
- GeMoMa makes the predictions for each reference transcript independently. Hence, it can occur that some of predictions of different reference transcripts overlap or are identical especially in gene families. Typically, you might like to filter or rank these predictions. We have implemented GAF (cf.
java -jar GeMoMa-<version>.jar CLI GAF) to do this automatically. However, you can also do it by hand using the GFF attributes. Using RNA-seq data in GeMoMa yields additional fields in the annotation that can be used, e.g., average coverage (avgCov). - A lot of transcripts have been filtered out by the Extractor. What can I do?
- There are several reasons for removing transcripts by the Extractor. At least in two cases you can try to get more transcripts by setting specific parameter values. First, if the transcript contains ambiguous nucleotides, please test the parameter "Ambiguity". Second, sometimes we received GFFs which contain wrong phases for CDS entries (e.g., 0 for all CDS entries in the phase column of the GFF). Since version 1.3.2, we provide the option "r" which stands for repair. If r=true is chosen, the Extractor tries to infer all phases for transcripts that show an error and would be filtered out.
- Is GeMoMa able to predict pseudo-genes/ncRNA?
- No, currently not.
- My RNA-seq data indicates there is an additional intron in a transcipt, but GeMoMa does not predict this. Or vice versa, GeMoMa predicts an intron that is not supported by RNA-seq data. What's the reason?
- GeMoMa is mainly based on the assumptions of amino acid and intron position conservation between reference and target species. Hence, GeMoMa tries to predict a gene model with similar exon-intron-structure in the target species and does not stick too much with RNA-seq data. Although intron position conservation can be observed in most cases, sometimes new introns evolve or others vanish. For this reasons, GeMoMa also allows for the inclusion or exclusion of introns adding some additional costs (cf. GeMoMa parameter intron-loss-gain-penalty). However, the behaviour of GeMoMa depends on the parameters settings (especially intron-loss-gain-penalty, sm (substitution matrix), go (gap opening), ge (gap extension)) and the length of the missed/additional intron. Nevertheless, such cases can only occur if the additional/missed intron has a length that can be divided by 3 preserving the reading frame.
- Since the available RNA-seq data only reflects a fraction of tissues/environmental conditions/..., missing RNA-seq evidence does not necessarily mean that the predictions is wrong.
- My RNA-seq data indicates two alternative, highly overlapping introns. Interestingly, GeMoMa does not take the intron that is more abundant. Why?
- GeMoMa reads the introns from the input file using some filter (cf. GeMoMa parameter r (reads)). All introns that pass the filter are used and treated equally. Hence, GeMoMa uses the intron that matches the expectation of intron position and amino acid conservation compared to the reference transcript.
Version history
GeMoMa 1.4 (03.05.2017)
- AnnotationEvidence: new tool computing tie and tpc for given annotation (gff)
- CompareTranscripts: new tool comparing predicted and given annotation (gff)
- Extractor:
- reading CDS with no parent tag (cf. discontinuous feature)
- automatic recognition of GFF or GTF annotation
- Warning if sequences mentioned in the annotation are not included in the reference sequence
- GeMoMa:
- allowing for multiple intron and coverage files (= using different library types at the same time)
- NA instead of "?" for tae, tde, tie, minSplitReads of single coding exon genes
- new default values for the parameters: predictions (10 instead of 1) and contig threshold (0.4 instead of 0.9)
- bugfix (write pc and minCov if possible for last CDS part in predicted annotation)
- bugfix (ref-gene name if no assignment is used)
- bugfix (minSplitReads, minCov, tpc, avgCov if no coverage available)
- GAF:
- nested genes on the same strand
- bugfix (if nothing passes the filter)
GeMoMa 1.3.2 (18.01.2017)
- Extractor: new parameter repair for broken transcript annotations
- GeMoMa: bugfixes (splice site computation)
GeMoMa 1.3.1 (09.12.2016)
- GeMoMa bugfix (finding start/stop codon for very small exons)
GeMoMa 1.3 (06.12.2016)
- ERE: new tool for extracting RNA-seq evidence
- Extractor: offers options for
- partial gene models
- ambiguities
- GeMoMa:
- RNA-seq
- defining splice sites
- additional feature in GFF and output
- transcript intron evidence (tie)
- transcript acceptor evidence (tae)
- transcript donor evidence (tde)
- transcript percentage coverage (tpc)
- ...
- improved GFF
- simplified the command line parameters
- IMPORTANT: parameter names changed for some parameters
- RNA-seq
- GAF: new tool for filtering and combining different predictions (especially of different reference organisms)
GeMoMa 1.1.3 (06.06.2016)
- minor modifications to the Extractor tool
GeMoMa 1.1.2 (05.02.2016)
- GeMoMa bugfix (upstream, downstream sequence for splice site detection)
- Extractor: new parameter s for selecting transcripts
- improved Galaxy integration
GeMoMa 1.1.1 (01.02.2016)
- initial release for paper